What is LoCOP DB: this database contains tens of thousands of gene pairs (protein coding and lincRNA genes) within 1Mb of each other that found to be significantly co-expressed across 49 GTEx (v8) human tissues. Moreover, genetic variants associated with the expression of both genes (expression Quantitative Trait Loci, eQTLs) are shown.
COP detection pipeline: Genome-wide maps of local gene co-expression were obtained from cross-individual gene expression quantifications (e.g. a gene expression matrix from multiple RNA-seq experiments), by exploiting the principle that nearby genes exhibiting expression correlation with each other across individuals, higher than what is expected by chance, can be considered as co-expressed. Briefly, the pipeline has the following steps:
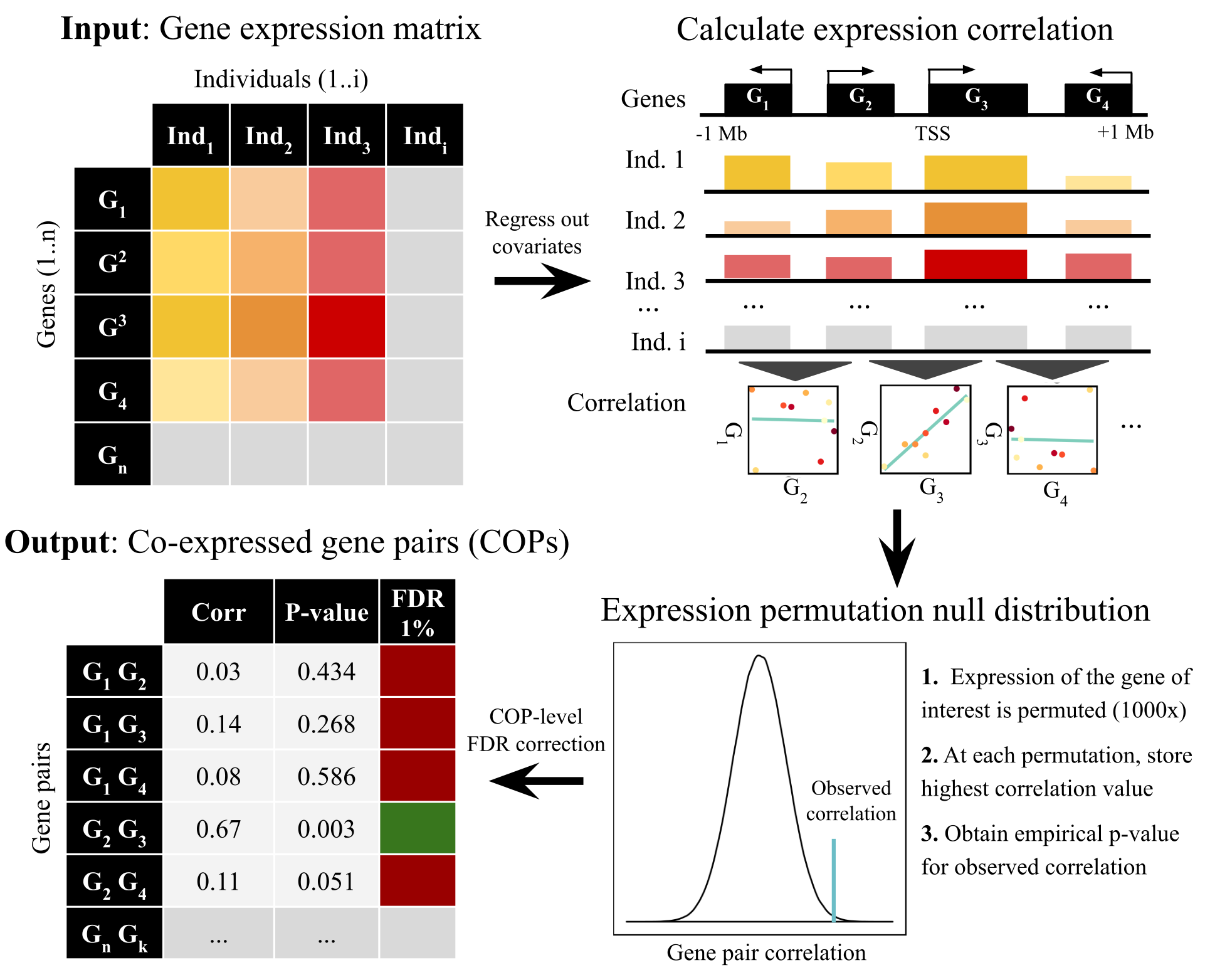
- Gene expression preparation: pre-filter (only protein-coding and lincRNAs were used) and normalise expression values. Extensively account for known (e.g. sex, subpopulation structure) and unknown confounding factors (using PEER covariates provided by GTEx) by regressing out each covariate from the gene expression matrix.
- For each gene, identify all genes in a cis window of 1Mb around the gene transcription start site (TSS) and calculate inter-individual expression correlation (Pearson correlation).
- For each gene/cis-gene pair, compare the observed values to expected correlation values under the null obtained by shuffling expression values and empirical p-values. This ensures that the number of nearby genes is accounted for.
- High observed correlation values are pinpointed and sets of bona fide co-expressed gene pairs (COPs) extracted by controlling for false discovery rate ( FDR 1% used here).
Note: Genes in non-autosomal chromosomes (e.g. X, Y, MT) as well as genes around the MHC region (chr6:29500000-33600000) were excluded. Gene models are based on Gencode v26 .
eQTL mapping: cis-eQTLs (within 1Mb) were mapped for each gene using QTLtools v1.1 cis function. To detect eQTLs affecting co-expressed genes, for each COP, the lead eQTL (FDR 5%) of each eGene was tested for their ability to associate (i.e. nominal p-value < 0.05, same effect sign) with the other co-expressed gene. This way, a COP may have 0, 1 or 2 associated shared eQTLs.
How to cite: Ribeiro DM, Rubinacci S, Ramisch A, Hofmeister RJ, Dermitzakis ET, Delaneau O. The molecular basis, genetic control and pleiotropic effects of local gene co-expression. Nat. Commun. 12, 4842 (2021).
Who are we: LoCOP DB is developed and maintained by the Systems and Population Genetics Group at the University of Lausanne, Switzerland. Website developed by Simone Rubinacci and Diogo Ribeiro. This work was funded by the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 885998.
Questions or suggestions: drop us an email at diogo.ribeiro [at] unil.ch



